Not a big fan of pandas dataframe syntax? Prefer to use SQL? Then suffer no more! In this detailed, step-by-step guide using RIPE Atlas public data, find out how DuckDB allows you to use SQL without any DB server on a Jupyter Notebook.
Say, which one of these two code snippets do you find more intelligible?
This Pandas Dataframe:
filtered_df = df[df['rdata'].str.match(r'b\d-[A-Za-z]{3}', na=False)]
result = filtered_df.groupby('rdata').agg({'rt': 'mean', 'rdata': 'size'}).rename(columns={'rdata': 'responses'}).reset_index()
Or this DuckDB syntax
res=con.sql('''
select rdata,avg(rt),count(1) as responses from atlas where
rdata ~ 'b[0-9]-[A-Za-z]{3}'
AND rdata IS NOT NULL
group by rdata
''')
If you like SQL more and are often confused or irritated by pandas dataframe syntax, suffer no more! You can use SQL instead without any DB server on a Jupyter Notebook. And it's fast.
Here, we want to showcase DuckDB, an open-source database that can be used for RIPE Atlas measurement analysis (or any data analysis).
DuckDB is:
- Fully integrated with Python, Jupyter, and Pandas (so you can import and export from and to)
- Allows SQL execution on data
- An in-memory database
- Severless
- Open-source
- More info: https://duckdb.org/docs/api/python/overview
Goals of this post
- Demonstrate how to use
duckdbas an alternative to Pandas for data analysis, featuring public RIPE Atlas data - Compare the syntax of
duckdbwith that of Pandas through simple examples - Provide helpful starting points for beginners exploring RIPE Atlas datasets
The inspiration for this post came from working with duckdb in a joint peer-reviewed paper and also from seeing that students at TU Delft were glad to switch to SQL for data analysis once they started using duckdb.
Pandas vs DuckDB
Part 1: Data preparation
#setting the measurements we will analyse
#b-root atlas measurements, `chaos queries`, 15min
# see more on https://www.isi.edu/~johnh/PAPERS/Moura16b.pdf for more on these measurements
b_root_measurements="https://atlas.ripe.net/api/v2/measurements/10310/results/?start=1730025000&stop=1730025900&format=json"
#Altas probes metadata, daily compiled
probes_file="https://ftp.ripe.net/ripe/atlas/probes/archive/2024/10/20241027.json.bz2"
#import python libraries
import pandas as pd
import json
import duckdb
import requests
import bz2
from io import BytesIO
Part 1a: Import RIPE Atlas measurements
We are going to present two ways of import JSON data to DuckDB:
- One-liner
- Step-by-step
You can pick each one you prefer - one liner is a bit more complex
One-liner import
In this complex SQL query, we do the following:
- Read all the measurements under data with a CTE
WITH data AS (SELECT unnest(result), *
FROM read_json($file_name)
)
- Read the JSON from RIPE Atlas measurements with
read_json($file_name) - Unnest the json object under result woth
unnest(result)
SELECT *, unnest(coalesce(answers, [NULL])) as ans
FROM data
- Use it in a FROM statement to extract the rdata
SELECT prb_id, timestamp, src_addr, rt, regexp_extract(ans.rdata, '\\[\"(.*)\"\\]', 1) as rdata FROM (
SELECT *, unnest(coalesce(answers, [NULL])) as ans
FROM data
)
- Package everything under
CREATE OR REPLACE TABLE atlas AS
Putting all this together, here's the query:
## one liner data import
#create a new database
con = duckdb.connect('ripe')
# SQL queries
# this query crates a table name atlas, then, gets the b_root_measurements JSON file and performs a select on it
# it may be too complex, so we provide an alternative version below
query = con.sql(
"""
CREATE OR REPLACE TABLE atlas AS (
WITH data AS (
SELECT unnest(result), *
FROM read_json($file_name)
)
SELECT prb_id, timestamp, src_addr, rt, regexp_extract(ans.rdata, '\\[\"(.*)\"\\]', 1) as rdata FROM (
SELECT *, unnest(coalesce(answers, [NULL])) as ans
FROM data
)
)
""",
params={
"file_name": b_root_measurements
}
)
row_count = con.sql("SELECT count() AS row_count FROM atlas")
row_count┌───────────┐
│ row_count │
│ int64 │
├───────────┤
│ 46016 │
└───────────┘
select_one = con.sql("SELECT * FROM atlas LIMIT 5")
select_one
┌─────────┬────────────┬────────────────┬────────┬─────────┐
│ prb_id │ timestamp │ src_addr │ rt │ rdata │
│ int64 │ int64 │ varchar │ double │ varchar │
├─────────┼────────────┼────────────────┼────────┼─────────┤
│ 1006359 │ 1730025150 │ 172.30.105.107 │ 26.725 │ b3-ams │
│ 1006359 │ 1730025389 │ 172.30.105.107 │ 24.782 │ b3-ams │
│ 1006359 │ 1730025631 │ 172.30.105.107 │ 28.913 │ b3-ams │
│ 1006359 │ 1730025871 │ 172.30.105.107 │ 25.327 │ b3-ams │
│ 1006154 │ 1730025174 │ 192.168.0.104 │ 4.15 │ b1-ams │
└─────────┴────────────┴────────────────┴────────┴─────────┘Alternative: Step-by-step data import
# step-by-step data import -- easier to undertand
b_root=requests.get(b_root_measurements)
b_root_data=b_root.json()
#sample data
b_root_data[0]
{'fw': 5080,
'mver': '2.6.2',
'lts': 57,
'dst_addr': '170.247.170.2',
'dst_port': '53',
'af': 4,
'src_addr': '172.30.105.107',
'proto': 'UDP',
'result': {'rt': 26.725,
'size': 50,
'abuf': '5iiAAAABAAEAAAAACGhvc3RuYW1lBGJpbmQAABAAA8AMABAAAwAAAAAABwZiMy1hbXM=',
'ID': 58920,
'ANCOUNT': 1,
'QDCOUNT': 1,
'NSCOUNT': 0,
'ARCOUNT': 0,
'answers': [{'TYPE': 'TXT', 'NAME': 'hostname.bind', 'RDATA': ['b3-ams']}]},
'msm_id': 10310,
'prb_id': 1006359,
'timestamp': 1730025150,
'msm_name': 'Tdig',
'from': '88.117.222.37',
'type': 'dns',
'stored_timestamp': 1730025220}
# define a method to parse the data from the JSON file
# so we can add to a list and later import into a dataframe and a duckdb table
def extract_info(data):
prb_id = data.get('prb_id')
timestamp = data.get('timestamp')
src_addr = data.get('src_addr')
rt = data.get('result', {}).get('rt')
answers = data.get('result', {}).get('answers', [{}])
# Ensure 'answers' is a non-empty list and 'RDATA' is in the first item
rdata = None
if answers and 'RDATA' in answers[0]:
rdata = answers[0]['RDATA'][0] if answers[0]['RDATA'] else 'None'
return {
'prb_id': prb_id,
'timestamp': timestamp,
'src_addr': src_addr,
'rt': rt,
'rdata': rdata
}
# now let' s parse the json and extract the fields we wish using extract_info
data=[]
for k in b_root_data:
data.append(extract_info(k))
print('We have retrieved ', len(data), ' measurements from Ripe Atlas')
We have retrieved 46,016 measurements from Ripe Atlas.
# now let's import it to a df
df = pd.DataFrame(data)
#create a new database
con=duckdb.connect('ripe')
# Drop the table if it exists, then create a new one and insert the DataFrame
# con.execute("DROP TABLE IF EXISTS atlas") -- Not needed, as DuckDB has CREATE OR REPLACE
con.execute("CREATE OR REPLACE TABLE df_atlas (prb_id INTEGER, timestamp INTEGER, src_addr TEXT, rt DOUBLE, rdata TEXT)")
con.execute("INSERT INTO df_atlas SELECT * FROM df")
result = con.sql("SELECT count() AS row_count FROM df_atlas").fetchone()
# Get the row count
row_count = result[0]
row_count
row_count_duckdb = con.sql("SELECT count(1) AS row_count FROM atlas")
row_count_duckdb
┌───────────┐
│ row_count │
│ int64 │
├───────────┤
│ 46016 │
└───────────┘## Sanity check
print("dataset len is:\n\t", len(data))
print("df len is:\n\t", len(df))
print('duckdb table len is\n',row_count)
print('duckdb table len is\n',row_count_duckdb)
dataset len is: 46016
df len is: 46016
duckdb table len is 46016
duckdb table len is:
┌───────────┐
│ row_count │
│ int64 │
├───────────┤
│ 46016 │
└───────────┘So, in summary: to import JSON DNS from Ripe Atlas, you can use both methods. Next we move to import RIPE Atlas metadata.
Part 1b: Import Atlas Probes metadata
Just like we did for the DNS JSON data (b_root_measurements), we will import this data in two ways:
- One-liner
- Step-by-step
One-liner import
To automatically read the compressed data with DuckDB and create a table with it, we will use the community extension shellfs. The DuckDB Python API allows us to install an extension and load it.
Note: The default repository for the extensions iscore. Asshellfsis part of the community extensions, we have to indicate the repository when installing the extension.
# Let's use DuckDB Community Extensions to get the archive and uncompress
# https://community-extensions.duckdb.org/extensions/shellfs.html
con.sql('INSTALL shellfs FROM community; ')
#if the above does not work you can try:
#con.install_extension("shellfs", repository="community")
con.load_extension("shellfs")
probes_data = con.sql(
"""
CREATE OR REPLACE TABLE probes AS (
SELECT *
FROM read_json("curl -s https://ftp.ripe.net/ripe/atlas/probes/archive/2024/10/20241027.json.bz2 | bunzip2 -d | jq '.objects' |")
)
"""
)
con.sql("SELECT * FROM probes LIMIT 5")
┌───────┬────────────────┬──────────────────────┬────────┬───┬──────────┬──────────────────────┬─────────────┐
│ id │ address_v4 │ address_v6 │ asn_v4 │ … │ day │ probe │ status_name │
│ int64 │ varchar │ varchar │ int64 │ │ varchar │ varchar │ varchar │
├───────┼────────────────┼──────────────────────┼────────┼───┼──────────┼──────────────────────┼─────────────┤
│ 1 │ 45.138.229.91 │ 2a10:3781:e22:1:22… │ 206238 │ … │ 20241027 │ https://atlas.ripe… │ Connected │
│ 2 │ NULL │ NULL │ 1136 │ … │ 20241027 │ https://atlas.ripe… │ Abandoned │
│ 3 │ 77.174.76.85 │ 2a02:a467:f500:1:2… │ 1136 │ … │ 20241027 │ https://atlas.ripe… │ Connected │
│ 4 │ 83.163.50.165 │ 2001:980:57a4:1:22… │ 3265 │ … │ 20241027 │ https://atlas.ripe… │ Abandoned │
│ 5 │ 83.163.239.181 │ 2001:981:602b:1:22… │ 3265 │ … │ 20241027 │ https://atlas.ripe… │ Abandoned │
├───────┴────────────────┴──────────────────────┴────────┴───┴──────────┴──────────────────────┴─────────────┤
│ 5 rows 20 columns (7 shown) │
└────────────────────────────────────────────────────────────────────────────────────────────────────────────┘#let's create a probe list to create later a dataframe and a new table
## now we're going to download the probes metadata file
#ok, let's now get the probes metadata
# Download the file
response = requests.get(probes_file)
compressed_data = BytesIO(response.content)
# Decompress the data in memory
with bz2.BZ2File(compressed_data, 'rb') as file:
probes = json.load(file)
with open("output.json", 'w') as file:
json.dump(probes['objects'], file)
#print sample data
print(probes['objects'][0])
pr_list=[]
for k in probes['objects']:
pr_list.append(k)
#pandas df of probes
df_prb=pd.DataFrame(pr_list)
# now let's create a duckdb table
# yeah, we use pandas for it :)
# Create a table and insert the DataFrame
con.execute("DROP TABLE IF EXISTS df_prb")
con.execute("CREATE TABLE IF NOT EXISTS probes AS SELECT * FROM df_prb")
# Verify the data in the DuckDB table
test_query = con.sql("SELECT count(1) AS row_count FROM probes").fetchone()
# Get the row count
row_count_probes = test_query[0]
row_count_probes = con.sql("SELECT count(1) AS row_count FROM probes").fetchone()
#sanity check
print("dataset len is:", len(probes['objects']))
print("df_prb len is:", len(df_prb))
print('duckdb table len is:',row_count_probes)
dataset len is: 42,365
df_prb len is 42,365
duckdb table len is: 42,365
Part 2: Data analysis
OK, so now we have two the same datasets (RIPE Atlas measurements and probes metadatata) stored in two different ways:
- Pandas dataframes:
- Measurements=
df - probes's metadata=
df_prb
- Measurements=
- Duckdb tables:
- Measurements=
atlas - probes's metadata=
probes
- Measurements=
Let's see how we can perform the same operations on both:
# let's start with how many queries we have
# let's compute the average RTT to each of the anycast server of b-root
# each site (each serfver location) is identified by the rdata field
# option 1: pandas
# Compute average rt for each rdata
average_rt = df.groupby('rdata')['rt'].mean().reset_index()
print(average_rt)
rdata rt
0 DNS1 0.724500
1 DNS2 2.280000
2 DNS3 1.031000
3 DNS_AF_NH3_01 4.687000
4 DNS_AF_NH3_13 4.401000
5 DNS_AF_NH3_15 4.119000
6 DNS_AF_NTH_16 4.109000
7 NS1 1.195000
8 OPNsense.local.lan 1.421500
9 OPNsense.stuyts.com 0.464750
10 OPNsense.zunie.lan 0.467750
11 alliance-45 1.789000
12 b 65.556000
13 b1-ams 36.861626
14 b1-iad 53.347860
15 b1-lax 122.322482
16 b1-mia 127.123686
17 b1-scl 25.029500
18 b1-sin 70.835360
19 b2-iad 54.859029
20 b2-lax 108.799266
21 b2-mia 127.405742
22 b2-scl 25.383507
23 b2-sin 72.801364
24 b3-ams 35.842288
25 b3-iad 58.851777
26 b3-lax 113.668726
27 b3-mia 114.486817
28 b3-scl 20.429663
29 b3-sin 80.070575
30 b4-ams 36.690085
31 b4-iad 51.383624
32 b4-lax 118.287110
33 b4-mia 135.760622
34 b4-sin 78.729474
35 cachedns.bosveld.co.za 1.117636
36 centralpi 1.205000
37 dur-01-dns-cluster 33.576250
38 elb-01-dns 35.105000
39 g2sanfelipe 5.373000
40 g3maracaibo 7.273500
41 gateway.bt.lan 0.381250
42 gw01.jacobsylvia.com 0.917750
43 localhost.localdomain 17.728750
44 netlan-1 1.184250
45 ns1 2.947000
46 ns2 0.755333
47 okctms2.boit.us 2.101000
48 opnsense.altus-escon.com 1.065500
49 opnsense.freebsd.vm.pve.myers.app 1.328000
50 pd3ajmeddx4li6ml.vla.yp-c.yandex.net 14.814750
51 proxy2 3.022500
52 res100.qmxp1.rrdns.pch.net 453.884333
53 res701.qmxp1.rrdns.pch.net 483.324000
54 thorin.mekom.ba 6.276250
55 tir-02-dns 120.030333
56 tns 0.618750
57 wall.obskure.net 1.410667Dealing with bogus data
As shown above, there are many results, but let's filtered out the valid ones. Some DNS queires from Ripe Atlas clients are intercepted and hijacked, see Moura16b and Nosyk23a about it. This means we need to filter out the data.
We can do it by looking at the rdata strings. For B-ROOT, it has the follow format:
bn-ABC, wherenis a integer andABCis a three letter string, denoting Airport Code
So let's see how can we do this filtering in both Pandas and Duckdb
# Filter rows where rdata matches the pattern bn-ABC, with Pandas
filtered_df = df[df['rdata'].str.match(r'b\d-[A-Za-z]{3}', na=False)]
# Compute average rt and count for each rdata
result = filtered_df.groupby('rdata').agg({'rt': 'mean', 'rdata': 'size'}).rename(columns={'rdata': 'responses'}).reset_index()
print(result)
rdata rt responses
0 b1-ams 36.861626 9645
1 b1-iad 53.347860 1645
2 b1-lax 122.322482 465
3 b1-mia 127.123686 497
4 b1-scl 25.029500 60
5 b1-sin 70.835360 780
6 b2-iad 54.859029 1717
7 b2-lax 108.799266 470
8 b2-mia 127.405742 528
9 b2-scl 25.383507 144
10 b2-sin 72.801364 758
11 b3-ams 35.842288 9659
12 b3-iad 58.851777 1961
13 b3-lax 113.668726 485
14 b3-mia 114.486817 491
15 b3-scl 20.429663 83
16 b3-sin 80.070575 737
17 b4-ams 36.690085 9602
18 b4-iad 51.383624 1848
19 b4-lax 118.287110 447
20 b4-mia 135.760622 556
21 b4-sin 78.729474 887#Ok, so how would you do it in duckdb
res=con.sql('''
select rdata,avg(rt),count(1) as responses from atlas where
rdata ~ 'b[0-9]-[A-Za-z]{3}'
AND rdata IS NOT NULL
group by rdata
''')
print(res)
┌─────────┬────────────────────┬───────────┐
│ rdata │ avg(rt) │ responses │
│ varchar │ double │ int64 │
├─────────┼────────────────────┼───────────┤
│ b1-ams │ 36.86162602384655 │ 9645 │
│ b1-iad │ 53.34785957446813 │ 1645 │
│ b3-mia │ 114.48681670061107 │ 491 │
│ b4-iad │ 51.38362391774902 │ 1848 │
│ b2-iad │ 54.85902912055905 │ 1717 │
│ b3-lax │ 113.66872577319587 │ 485 │
│ b1-sin │ 70.83536025641033 │ 780 │
│ b4-lax │ 118.28710961968663 │ 447 │
│ b1-scl │ 25.02949999999999 │ 60 │
│ b2-scl │ 25.38350694444445 │ 144 │
│ b3-scl │ 20.429662650602403 │ 83 │
│ b3-ams │ 35.84228822859509 │ 9659 │
│ b1-mia │ 127.12368611670024 │ 497 │
│ b2-lax │ 108.79926595744679 │ 470 │
│ b1-lax │ 122.32248172043013 │ 465 │
│ b2-sin │ 72.80136411609497 │ 758 │
│ b2-mia │ 127.40574242424246 │ 528 │
│ b4-ams │ 36.69008508644033 │ 9602 │
│ b3-iad │ 58.85177715451294 │ 1961 │
│ b4-mia │ 135.76062230215834 │ 556 │
│ b3-sin │ 80.0705753052917 │ 737 │
│ b4-sin │ 78.7294735062007 │ 887 │
├─────────┴────────────────────┴───────────┤
│ 22 rows 3 columns │
└──────────────────────────────────────────┘I personally find the SQL syntax more readable. It's a matter of taste!
Part 3: Aggregation and visualisation
Shifting gears, the exercise now is to obtain the average RTT from the probes to B-ROOT, based on its country of origin. Note, however, that the country of origin is not on the results table, but on the probe metadata.
We need to join the results of two tables, so let's start with DuckDB:
''' We can do like this on SQL
* We join the tables on r.prb_id = p.id, which is the Atlas probe ID
* We filter probes that reach B-ROOT servers by looking into `rdata` string
* We group by country_code
'''
res = con.sql('''
SELECT
p.country_code,
AVG(r.rt) AS avg_rt,
COUNT(r.rt) AS total_measurements,
COUNT(DISTINCT r.prb_id) AS number_atlas_probes
FROM
atlas r
JOIN
probes p
ON
r.prb_id = p.id
WHERE
r.rdata ~ 'b[0-9]-[A-Za-z]{3}' AND r.rdata IS NOT NULL
GROUP BY
p.country_code
ORDER BY
avg_rt DESC;
''')
print(res)
┌──────────────┬────────────────────┬────────────────────┬─────────────────────┐
│ country_code │ avg_rt │ total_measurements │ number_atlas_probes │
│ varchar │ double │ int64 │ int64 │
├──────────────┼────────────────────┼────────────────────┼─────────────────────┤
│ VU │ 325.6395 │ 4 │ 1 │
│ NC │ 290.83149999999995 │ 16 │ 4 │
│ KI │ 272.45685714285713 │ 7 │ 2 │
│ MM │ 263.70525 │ 4 │ 1 │
│ NP │ 247.75570454545456 │ 44 │ 12 │
│ PK │ 225.83114285714288 │ 7 │ 2 │
│ CI │ 210.426 │ 1 │ 1 │
│ MW │ 207.539875 │ 8 │ 2 │
│ GQ │ 206.79975 │ 4 │ 1 │
│ CD │ 203.2782857142857 │ 14 │ 4 │
│ · │ · │ · │ · │
│ · │ · │ · │ · │
│ · │ · │ · │ · │
│ GB │ 24.116230922693237 │ 2005 │ 532 │
│ DK │ 23.834773269689745 │ 419 │ 111 │
│ CH │ 22.044400519031118 │ 1156 │ 310 │
│ LI │ 20.944277777777774 │ 18 │ 5 │
│ DE │ 20.81939130434787 │ 6003 │ 1636 │
│ GG │ 19.163 │ 4 │ 1 │
│ BE │ 16.389434494195683 │ 603 │ 161 │
│ LU │ 13.248532467532467 │ 154 │ 41 │
│ NL │ 12.010846872082148 │ 2142 │ 575 │
│ NULL │ 5.9505 │ 4 │ 1 │
├──────────────┴────────────────────┴────────────────────┴─────────────────────┤
│ 176 rows (20 shown) 4 columns │
└──────────────────────────────────────────────────────────────────────────────┘# now, let's do the same for pandas
# Merge DataFrames on prb_id and id
merged_df = pd.merge(df, df_prb, left_on='prb_id', right_on='id')
# Filter rows where rdata matches the pattern bX-ABC and is not None
filtered_df = merged_df[merged_df['rdata'].notna() & merged_df['rdata'].str.match(r'b[0-9]-[A-Za-z]{3}')]
# Group by country_code and calculate the required metrics
result = filtered_df.groupby('country_code').agg(
avg_rt=('rt', 'mean'),
total_measurements=('rt', 'size'),
number_atlas_probes=('prb_id', 'nunique')
).reset_index()
# Sort by avg_rt descending
result = result.sort_values(by='avg_rt', ascending=False)
print(result)
country_code avg_rt total_measurements number_atlas_probes
171 VU 325.639500 4 1
115 NC 290.831500 16 4
82 KI 272.456857 7 2
104 MM 263.705250 4 1
120 NP 247.755705 44 12
.. ... ... ... ...
40 DE 20.819391 6003 1636
55 GG 19.163000 4 1
14 BE 16.389434 603 161
95 LU 13.248532 154 41
118 NL 12.010847 2142 575
[175 rows x 4 columns]Again, I just prefer to write SQL to do these sort of queries and complex queries.
Sanity check
#sanity check
test_pandas=result[result['country_code']=='ES']
print("results pandas" )
print(test_pandas.to_string(index=False))
test_duckdb=con.sql('''
SELECT
p.country_code,
AVG(r.rt) AS avg_rt,
COUNT(r.rt) AS total_measurements,
COUNT(DISTINCT r.prb_id) AS number_atlas_probes
FROM
atlas r
JOIN
probes p
ON
r.prb_id = p.id
WHERE
r.rdata ~ 'b[0-9]-[A-Za-z]{3}' AND r.rdata IS NOT NULL
GROUP BY
p.country_code
HAVING
p.country_code = 'ES'
ORDER BY
avg_rt DESC;
''')
print("results duckdb")
print(test_duckdb.to_df().to_string(index=False))
results pandas
country_code avg_rt total_measurements number_atlas_probes
ES 42.226138 836 222
results duckdb
country_code avg_rt total_measurements number_atlas_probes
ES 42.226138 836 222#1. You can export your sql results to Dataframes and carry on
res = con.sql('''
SELECT
p.country_code,
AVG(r.rt) AS avg_rt,
COUNT(r.rt) AS total_measurements,
COUNT(DISTINCT r.prb_id) AS number_atlas_probes
FROM
atlas r
JOIN
probes p
ON
r.prb_id = p.id
WHERE
r.rdata ~ 'b[0-9]-[A-Za-z]{3}' AND r.rdata IS NOT NULL
GROUP BY
p.country_code
ORDER BY
avg_rt DESC;
''')
test_df=res.to_df()
print(test_df)
country_code avg_rt total_measurements number_atlas_probes
0 VU 325.639500 4 1
1 NC 290.831500 16 4
2 KI 272.456857 7 2
3 MM 263.705250 4 1
4 NP 247.755705 44 12
.. ... ... ... ...
171 GG 19.163000 4 1
172 BE 16.389434 603 161
173 LU 13.248532 154 41
174 NL 12.010847 2142 575
175 None 5.950500 4 1
[176 rows x 4 columns]# extra 2, some nice visualizations
# let's make a world map, heatmap
import geopandas as gpd
import matplotlib.pyplot as plt
import matplotlib.colors as colors
import numpy as np
from mpl_toolkits.axes_grid1 import make_axes_locatable
Now download some files to make some world map:
- Go to https://www.naturalearthdata.com/downloads/110m-cultural-vectors/
- Download
Admin 0 – Countries - Extract all files somewhere (I did
/tmp/for this post)
DuckDB with Geodata
For the geodata you can check out this extension: https://duckdb.org/docs/extensions/spatial/overview. Also, there are a lot of materials online about handling geodata with DuckDB.
import pandas as pd
import geopandas as gpd
import matplotlib.pyplot as plt
from mpl_toolkits.axes_grid1 import make_axes_locatable
from matplotlib import colors
# Load world map data from the downloaded shapefile
world = gpd.read_file("/tmp/ne_110m_admin_0_countries/ne_110m_admin_0_countries.shp") # Update this path
# Set the parameters to use Type 1 fonts
plt.rcParams['ps.useafm'] = True
plt.rcParams['pdf.use14corefonts'] = True
plt.rcParams['text.usetex'] = True
plt.rcParams['pdf.fonttype'] = 42
plt.rcParams["figure.autolayout"] = True
plt.rcParams.update({'font.size': 14})
# Merge the world GeoDataFrame with your DataFrame
world = world.merge(test_df, how="left", left_on="ISO_A2_EH", right_on="country_code")
# Create a new column for avg_rt values
world['avg_rt'] = world['avg_rt'].fillna(float('nan')) # Explicitly keep NaN for countries without data
# Define the legend labels (bins)
legend_labels = [0, 10, 30, 50, 80, 100, 150, 500] # Include 0 to handle cases without data
# Define intervals for the color mapping
world['legend_intervals'] = pd.cut(world['avg_rt'], bins=legend_labels, include_lowest=True, labels=False)
# Set colors for regions with no data
# We'll use a separate color for NaN values (e.g., light grey)
cmap = plt.get_cmap('RdYlGn_r', len(legend_labels) - 1)
cmap.set_under('lightgrey') # Set a color for regions with no data
# Plotting
fig, ax = plt.subplots(1, 1, figsize=(15, 10))
world.boundary.plot(ax=ax, linewidth=1)
# Plot the world map with the data, setting NaN to be transparent
world.plot(column='legend_intervals', cmap=cmap, linewidth=0.8, ax=ax, edgecolor='0.8',
missing_kwds={"color": "lightgrey", "label": "No Data"})
# Create a new axes for the color bar
divider = make_axes_locatable(ax)
cax = divider.append_axes("right", size="3%", pad=0.2)
# Create a custom color bar
norm = colors.BoundaryNorm(legend_labels, cmap.N)
sm = plt.cm.ScalarMappable(cmap=cmap, norm=norm)
sm.set_array([])
# Remove the coordinates from the axes
ax.axis('off')
# Add the custom color bar
fig.colorbar(sm, ticks=legend_labels, orientation='vertical', shrink=0.3, cax=cax)
# Color the oceans in light blue
ax.set_facecolor('lightblue')
# Optionally color the boundaries again if needed
world.boundary.plot(ax=ax, linewidth=0.5, color='black')
plt.title('Avg RTT (ms) to B-Root From Ripe Atlas Probes (2024-10-27 10:30:00 -- 11:00:00 UTC)\n[Countries in gray have no data]',
loc='center',fontsize=20, fontweight='bold', x=-15.8)
plt.show()
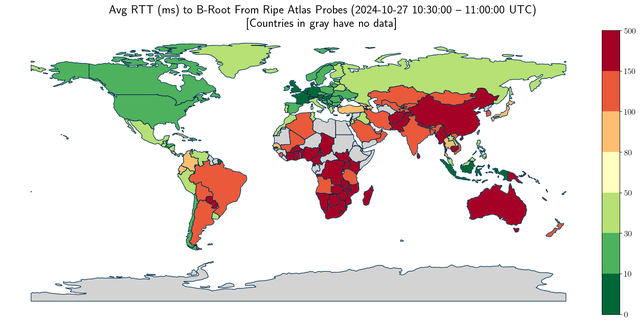
#now some graph sanity check
cc=['BR', 'US', 'ZA', 'JP', 'RU', 'AU', 'IN', 'NL', 'CL']
print('test_df country_code, test_df avg_rt, world ISO_A2_EH, world avg_rt')
for k in cc:
print(test_df[test_df['country_code']==k]['country_code'].values[0],
test_df[test_df['country_code']==k]['avg_rt'].values[0],
world[world['ISO_A2_EH']==k]['ISO_A2_EH'].values[0],
world[world['ISO_A2_EH']==k]['avg_rt'].values[0]
)
test_df country_code, test_df avg_rt, world ISO_A2_EH, world avg_rt
BR 129.26810119047633 BR 129.26810119047633
US 44.494604962139476 US 44.494604962139476
ZA 175.8397155425219 ZA 175.8397155425219
JP 89.54156737588649 JP 89.54156737588649
RU 70.50730801435407 RU 70.50730801435407
AU 180.4477780548629 AU 180.4477780548629
IN 115.01301185770751 IN 115.01301185770751
NL 12.010846872082148 NL 12.010846872082148
CL 34.110152263374495 CL 34.110152263374495Conclusion
- There's an alternative to dataframe language.
- DuckDB enables you to write clear SQL.
- We have shown an alternative engine for analysis for improved clarity.





Comments 1
The comments section is closed for articles published more than a year ago. If you'd like to inform us of any issues, please contact us.
Ties de Kock •
Nice article! duckdb has quickly become one of the favourite tools for my data analysis workflows. Since it turns out that many patterns that I end up doing in dataframes (split -> aggregate) are manageable in SQL. I even found some use case for python UDFs that I then call from SQL. And I like how easy it is to integrate it into workbooks or scripts and scales from small (megabytes) to medium-sized data (~500GB uncompressed dataframes). I have done some experiments with parquet-based derivative data of internet data. When they stabilise I will try to write more about it. So far, the only guarantee they come with is that I will change the format 😬